-Search query
-Search result
Showing 1 - 50 of 83 items for (author: yen & yc)

EMDB-39126: 
Structure of the FADD/Caspase-8/cFLIP death effector domain assembly
Method: single particle / : Lin SC, Yang CY

EMDB-39127: 
Structure of the FADD/Caspase-8/cFLIP death effector domain assembly
Method: single particle / : Lin SC, Yang CY
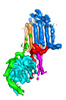
EMDB-40572: 
Human adenylyl Cyclase 5 in complex with Gbg
Method: single particle / : Yen YC, Tesmer JJG

EMDB-40573: 
Dimeric form of human adenylyl cyclase 5
Method: single particle / : Yen YC, Tesmer JJG

EMDB-40650: 
Phosphoinositide phosphate 3 kinase gamma
Method: single particle / : Chen CL, Tesmer JJG, Bandekar SJ, Cash J

EMDB-40651: 
Phosphoinositide phosphate 3 kinase gamma bound with ATP
Method: single particle / : Chen CL, Tesmer JJG, Bandekar SJ, Cash J

EMDB-40652: 
Phosphoinositide phosphate 3 kinase gamma bound with ADP
Method: single particle / : Chen CL, Tesmer JJG, Bandekar SJ, Cash J

EMDB-40653: 
Phosphoinositide phosphate 3 kinase gamma bound with ADP and Gbetagamma
Method: single particle / : Chen CL, Tesmer JJG, Bandekar SJ, Cash J

EMDB-40654: 
Phosphoinositide phosphate 3 kinase gamma bound with ADP and two Gbetagamma subunits in State 1
Method: single particle / : Chen CL, Tesmer JJG, Bandekar SJ, Cash J

EMDB-40655: 
Phosphoinositide phosphate 3 kinase gamma bound with ADP and two Gbetagamma subunits in State 2
Method: single particle / : Chen CL, Tesmer JJG, Bandekar SJ, Cash J

EMDB-41837: 
The structure of the PP2A-B56Delta holoenzyme mutant - E197K
Method: single particle / : Wu CG, Xing Y

EMDB-42018: 
The structure of the PP2A-B56Delta holoenzyme mutant - E197K
Method: single particle / : Wu CG, Xing Y

EMDB-34030: 
Cryo-EM map of a dimeric form of Ecoli Malate Synthase G (MSG)
Method: single particle / : Wu KP, Wu YM, Lu YC

EMDB-34029: 
2.9-angstrom cryo-EM structure of Ecoli malate synthase G
Method: single particle / : Wu KP, Wu YM, Lu YC

EMDB-33374: 
Focused refinement cryo-EM map of the A/B/C subunits of the T=4 lake sinai virus 2 virus-like particle at pH 7.5
Method: single particle / : Chen NC, Wang CH, Chen CJ, Yoshimura M, Guan HH, Chuankhayan P, Lin CC

EMDB-33375: 
Focus refinement cryo-EM map of the D/D/D subunits of the T=4 lake sinai virus 2 virus-like particle
Method: single particle / : Chen NC, Wang CH, Chen CJ, Yoshimura M, Guan HH, Chuankhayan P, Lin CC

EMDB-33376: 
Focused refinement cryo-EM map of the A/B/C subunits of the T=3 lake sinai virus 2 virus-like particle at pH 7.5
Method: single particle / : Chen NC, Wang CH, Chen CJ, Yoshimura M, Guan HH, Chuankhayan P, Lin CC

EMDB-33377: 
Focused refinement cryo-EM map of the A/B/C subunits of the T=4 lake sinai virus 2 virus-like particle at pH 6.5
Method: single particle / : Chen NC, Wang CH, Chen CJ, Yoshimura M, Guan HH, Chuankhayan P, Lin CC

EMDB-33378: 
Focused refinement cryo-EM map of the D/D/D subunits of the T=4 lake sinai virus 2 virus-like particle at pH 6.5
Method: single particle / : Chen NC, Wang CH, Chen CJ, Yoshimura M, Guan HH, Chuankhayan P, Lin CC

EMDB-33379: 
Focused refinement cryo-EM map of the A/B/C subunits of the T=3 lake sinai virus 2 virus-like particle at pH 6.5
Method: single particle / : Chen NC, Wang CH, Chen CJ, Yoshimura M, Guan HH, Chuankhayan P, Lin CC

EMDB-33380: 
Focused refinement cryo-EM map of the A/B/C subunits of the T=4 lake sinai virus 2 virus-like particle at pH 8.5
Method: single particle / : Chen NC, Wang CH, Chen CJ, Yoshimura M, Guan HH, Chuankhayan P, Lin CC

EMDB-33381: 
Focused refinement cryo-EM map of the D/D/D subunits of the T=4 lake sinai virus 2 virus-like particle at pH 8.5
Method: single particle / : Chen NC, Wang CH, Chen CJ, Yoshimura M, Guan HH, Chuankhayan P, Lin CC

EMDB-33382: 
Focused refinement cryo-EM map of the A/B/C subunits of the T=3 lake sinai virus 2 virus-like particle at pH 8.5
Method: single particle / : Chen NC, Wang CH, Chen CJ, Yoshimura M, Guan HH, Chuankhayan P, Lin CC

EMDB-33383: 
Focused refinement cryo-EM map of the A/B/C subunits of the T=3 lake sinai virus 1 (delta N-terminal 48 residues) virus-like particle at pH 6.5
Method: single particle / : Chen NC, Wang CH, Chen CJ, Yoshimura M, Guan HH, Chuankhayan P, Lin CC

EMDB-33384: 
Cryo-EM map of the T=4 lake sinai virus 1 (delta N-terminal 48 residues) virus-like particle at pH 6.5
Method: single particle / : Chen NC, Wang CH, Chen CJ, Yoshimura M, Guan HH, Chuankhayan P, Lin CC

EMDB-33368: 
Cryo-EM structure of the T=3 lake sinai virus 2 virus-like capsid at pH 7.5
Method: single particle / : Chen NC, Wang CH, Chen CJ, Yoshimura M, Guan HH, Chuankhayan P, Lin CC

EMDB-33369: 
Cryo-EM structure of the T=4 lake sinai virus 2 virus-like capsid at pH 6.5
Method: single particle / : Chen NC, Wang CH, Chen CJ, Yoshimura M, Guan HH, Chuankhayan P, Lin CC

EMDB-33370: 
Cryo-EM structure of the T=3 lake sinai virus 2 virus-like capsid at pH 6.5
Method: single particle / : Chen NC, Wang CH, Chen CJ, Yoshimura M, Guan HH, Chuankhayan P, Lin CC

EMDB-33371: 
Cryo-EM structure of the T=4 lake sinai virus 2 virus-like capsid at pH 8.5
Method: single particle / : Chen NC, Wang CH, Chen CJ, Yoshimura M, Guan HH, Chuankhayan P, Lin CC

EMDB-33372: 
Cryo-EM structure of the T=3 lake sinai virus 2 virus-like capsid at pH 8.5
Method: single particle / : Chen NC, Wang CH, Chen CJ, Yoshimura M, Guan HH, Chuankhayan P, Lin CC

EMDB-33373: 
Cryo-EM structure of the T=3 lake sinai virus 1 (delta-N48) virus-like capsid at pH 6.5
Method: single particle / : Chen NC, Wang CH, Chen CJ, Yoshimura M, Guan HH, Chuankhayan P, Lin CC

EMDB-34806: 
SARS-CoV-2 Delta Spike in complex with FP-12A
Method: single particle / : Chen X, Wu YM

EMDB-34807: 
SARS-CoV-2 Delta Spike in complex with IS-9A
Method: single particle / : Mohapatra A, Wu YM

EMDB-34808: 
SARS-CoV-2 Omicron BA.1 Spike in complex with IY-2A
Method: single particle / : Chen X, Mohapatra A, Wu YM

EMDB-32329: 
Cryo-EM map of PEDV (Pintung 52) S protein with all three protomers in the D0-down conformation determined in situ on intact viral particles.
Method: single particle / : Hsu STD, Draczkowski P, Wang YS

EMDB-32332: 
Subtomogram averaging of PEDV (Pintung 52) S protein with all three protomers in the D0-down conformation determined in situ on intact viral particles.
Method: subtomogram averaging / : Hsu STD, Draczkowski P, Wang YS, Huang CY

EMDB-32333: 
Subtomogram averaging of PEDV (Pintung 52) S protein with one protomer in the D0-up conformation and two protomers in the D0-down conformation, determined in situ on intact viral particles
Method: subtomogram averaging / : Hsu STD, Draczkowski P, Wang YS, Huang CY

EMDB-32337: 
Subtomogram averaging of PEDV (Pintung 52) S protein with two protomers in the D0-up conformation and one protomer in the D0-down conformation, determined in situ on intact viral particles.
Method: subtomogram averaging / : Hsu STD, Draczkowski P, Wang YS, Huang CY

EMDB-32338: 
Cryo-EM map of PEDV S protein with one protomer in the D0-up conformation while the other two in the D0-down conformation
Method: single particle / : Hsu STD, Draczkowski P, Wang YS

EMDB-32339: 
Subtomogram averaging of PEDV (Pintung 52) S protein with all three protomers in the D0-up conformation determined in situ on intact viral particles.
Method: subtomogram averaging / : Hsu STD, Draczkowski P, Wang YS, Huang CY

EMDB-32340: 
Subtomogram averaging of PEDV (Pintung 52) S protein in the postfusion form determined in situ on intact viral particles.
Method: subtomogram averaging / : Hsu STD, Draczkowski P, Wang YS, Huang CY

EMDB-33646: 
Cryo-EM map of IPEC-J2 cell-derived PEDV PT52 S protein with three D0-up
Method: single particle / : Hsu STD, Draczkowski P, Wang YS

EMDB-33647: 
Cryo-EM map of IPEC-J2 cell-derived PEDV PT52 S protein one D0-down and two D0-up
Method: single particle / : Hsu STD, Draczkowski P, Wang YS

EMDB-33648: 
Symmetry-expanded and locally refined protomer structure of IPEC-J2 cell-derived PEDV PT52 S with a CTD-close conformation
Method: single particle / : Hsu STD, Draczkowski P, Wang YS
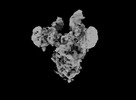
EMDB-33649: 
Symmetry-expanded and locally refined protomer structure of IPEC-J2 cell-derived PEDV PT52 S with a CTD-open conformation
Method: single particle / : Hsu STD, Draczkowski P, Wang YS

EMDB-33700: 
Cryo-EM map of HEK293F cell-derived PEDV PT52 S protein with three D0-down
Method: single particle / : Hsu STD, Draczkowski P, Wang YS

EMDB-33701: 
Cryo-EM map of HEK293F cell-derived PEDV PT52 S protein one D0-up and two D0-down
Method: single particle / : Hsu STD, Draczkowski P, Wang YS

EMDB-33702: 
Cryo-EM map of HEK293F cell-derived PEDV PT52 S protein with three D0-up
Method: single particle / : Hsu STD, Draczkowski P, Wang YS

EMDB-33703: 
Cryo-EM map of HEK293F cell-derived PEDV PT52 S T326I with three D0-down
Method: single particle / : Hsu STD, Draczkowski P, Wang YS

EMDB-33704: 
Cryo-EM map of HEK293F cell-derived PEDV PT52 S T326I one D0-up and two D0-down
Method: single particle / : Hsu STD, Draczkowski P, Wang YS
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
